Author
Author  Correspondence author
Correspondence author
Cancer Genetics and Epigenetics, 2024, Vol. 12, No. 3
Received: 10 May, 2024 Accepted: 12 Jun., 2024 Published: 27 Jun., 2024
The application of cancer mutation analysis in drug development offers new opportunities for personalized treatment, better meeting patient needs while reducing unnecessary treatments and potential side effects. Additionally, it revolutionizes drug development by accelerating the research and approval of new drugs. With the resolution of data management and privacy issues, the strengthening of interdisciplinary collaboration, and continuous technological advancements, cancer mutation analysis will continue to play a critical role in the future. This study aims to explore the key role of cancer mutation analysis in drug development, summarizing its methods and applications. It elaborates on genomic sequencing technologies and bioinformatics tools and databases, highlighting the importance of cancer mutation analysis in clinical applications. The goal is to promote the broad application of cancer mutation analysis, providing more effective treatment options for patients.
Cancer is a serious health issue that is widespread globally. According to the World Health Organization, cancer is one of the leading causes of death worldwide (Mattiuzzi and Lippi, 2019). Each year, millions of people are diagnosed with various types of cancer, and the incidence of cancer is on the rise. Cancer imposes a significant physical and psychological burden on patients and has a substantial impact on their families and society. Therefore, understanding the epidemiological characteristics of cancer is crucial for developing prevention and treatment strategies.
Despite significant medical advances in recent decades, cancer treatment remains a formidable challenge. Traditional treatments, such as radiotherapy and chemotherapy, are effective for certain types of cancer but have limited efficacy for others. These treatments often come with adverse reactions and side effects, causing suffering for patients. Additionally, drug resistance and tumor recurrence complicate cancer treatment further. Therefore, new methods and strategies are urgently needed to improve the effectiveness of cancer therapy.
Different types of cancer have distinct molecular characteristics, necessitating personalized treatment approaches to address specific situations. Global epidemiological studies of cancer have revealed variations in cancer incidence across different geographic regions and population groups (Liu et al., 2021), providing essential information for developing region-specific cancer prevention and control strategies. Mutation analysis is a critical component of personalized medicine and precision medicine. By gaining a deep understanding of mutations in cancer-related genes, we can support the development of more targeted treatment methods.
The purpose of this review is to explore the application of cancer mutation analysis in drug development. By gaining a deeper understanding of the classification, characteristics, and clinical applications of cancer mutations, this review focuses on gene mutations and mutation analysis technologies and how they influence cancer treatment and drug development. The review emphasizes the importance of cancer mutation analysis in formulating personalized treatment plans and advancing the field of drug development. It aims to provide an in-depth understanding of the latest advances and future potential of cancer mutation analysis in drug development to promote the development of more effective cancer treatment methods.
1 Classification and Characteristics of Cancer Mutations
1.1 Types of gene mutations
1.1.1 Point mutations
The classification and characteristics of cancer mutations are key topics in cancer research. Understanding these mutations helps to better comprehend tumor development and design relevant treatment methods. Point mutations are one of the most common types of gene mutations. They refer to changes in a single nucleotide within a gene (Parhami et al., 2020) and can be categorized into subtypes such as missense mutations, nonsense mutations, nucleotide insertions or deletions, and silent mutations.
Missense mutations are point mutations that result in the replacement of one amino acid with another (Figure 1). This type of mutation may alter the structure and function of proteins, thereby affecting normal cellular biological activities. Some missense mutations are considered oncogenic because they can activate or enhance the function of oncogenes. Nonsense mutations refer to a codon encoding an amino acid being replaced with a stop codon, leading to premature termination of protein synthesis. This usually results in the loss or defect of the protein, affecting cellular function.
.png) Figure 1 New ICT based fertility management model in private dairy farm India as well as abroad |
Nucleotide insertions or deletions involve the insertion or deletion of one or more nucleotides in a gene. This can cause a frameshift mutation, altering the amino acid sequence of the protein. Silent mutations refer to mutations that do not change the amino acid sequence because multiple codons encode the same amino acid. Although they do not alter the chemical properties of amino acids, they may still impact protein structure and function.
1.1.2 Deletion and insertion mutations
Deletion and insertion mutations are common types of gene mutations in cancer research that can significantly impact the normal biological functions of cells. Deletion mutations refer to the loss of some bases in the gene sequence, leading to changes in the DNA sequence. This may cause a frameshift mutation, altering the amino acid sequence of the protein. Deletion mutations often result in highly unstable proteins because they may generate new codons after the deleted region, potentially causing abnormal protein synthesis. In cancer, this type of mutation typically leads to the abnormal expression of key regulatory proteins in cancer cells, thereby promoting tumor growth and spread.
Insertion mutations involve the addition of extra bases into the gene sequence, resulting in changes to the DNA sequence. These extra bases can cause a frameshift mutation, altering the way proteins are synthesized. Similar to deletion mutations, insertion mutations can also lead to the production of abnormal proteins. These abnormal proteins may lose normal function or have enhanced oncogenic properties. Insertion mutations play a crucial role in some cancers, particularly in DNA repair genes, as these mutations can disrupt DNA damage repair mechanisms, increasing the susceptibility of cancer cells.
1.1.3 Structural variations
Structural variations play a key role in the occurrence and development of cancer because they can alter genetic information and gene expression patterns in cells. Structural variations involve large-scale changes in the DNA sequence, including gene rearrangements, chromosomal deletions, chromosomal duplications, and inversions. These variations can significantly impact genomic stability and normal cellular functions, thereby promoting tumor development. Understanding these variations helps researchers gain better insights into the molecular mechanisms of cancer.
Gene rearrangements refer to the reorganization of two different parts of a gene at the DNA level, typically involving chromosomal breaks and reattachments. This variation may produce fusion proteins, which can have oncogenic properties because they may lose normal regulation and promote uncontrolled cell growth. Chromosomal deletions involve the loss of a part or entire chromosome, which can lead to the inactivation of important suppressor genes, thereby increasing cancer risk. Chromosomal deletions can also cause chromosomal imbalance, affecting normal cellular functions.
Chromosomal duplications refer to the duplication of a part or entire chromosome. This variation may lead to the overexpression of certain genes, promoting tumor growth. Inversions involve the reversal of two parts of a chromosome. This variation can affect the structure and function of the chromosome, impacting normal cell growth and division.
1.2 Functional impacts of gene mutations
Gene mutations are one of the critical drivers of cancer development. They can be categorized into two main types based on their functional impact: activating mutations and inhibitory mutations. Both types of mutations can cause cells to lose normal growth and division control, promoting tumor formation. Studying the functional impacts of these gene mutations helps to better understand the mechanisms of cancer development and provides important information for developing therapeutic strategies.
1.2.1 Activating mutations
Activating mutations are mutations that enhance or increase the function of a gene. These mutations typically lead to the production of abnormal proteins that become overly active within the cell, promoting cancer development. Activating mutations often result in certain genes becoming oncogenes, which normally regulate cell growth and division. For example, activating mutations in the RAS family genes can lead to abnormal cell growth and division (Hodge et al., 2020), which is common in various cancers.
Activating mutations can also lead to the overactivation of signaling pathways that normally control cell growth and division. For instance, activating mutations can cause signaling proteins within the cell to become overly active, promoting uncontrolled cell growth. Additionally, activating mutations can result in the loss of normal gene expression regulation, including the loss of control by inhibitory factors, leading to the overexpression of certain genes within the cell. Activating mutations can also make cells resistant to apoptosis, a normal mechanism of cell death. This means that cancer cells can continue to grow and spread without being limited by the body's normal control mechanisms.
1.2.2 Inhibitory mutations
Inhibitory mutations are mutations that reduce or abolish the normal function of a gene, typically leading to the inhibition of cell growth and division. These mutations also play a significant role in cancer development. Inhibitory mutations can cause a gene to completely lose its normal function, meaning the gene can no longer perform its biological functions, such as regulating cell growth or repairing DNA. This can result in abnormal cell proliferation and uncontrolled growth.
Inhibitory mutations can also cause cells to lose their inhibitory effect on other genes. Normally, certain genes inhibit cell growth and division, but inhibitory mutations can cause this inhibition to fail, promoting abnormal cell proliferation. Some inhibitory mutations may lead to the failure of the cell's DNA repair mechanisms, meaning the cell cannot repair its own DNA damage. This can result in the accumulation of other mutations in the DNA, ultimately promoting cancer development. Inhibitory mutations can also cause cells to lose regulation of apoptosis (programmed cell death). This means that cells cannot undergo self-destruction, even if they have suffered severe DNA damage, and will continue to grow and spread.
2 Cancer Mutation Analysis Techniques
2.1 Genomic sequencing technologies
Cancer mutation analysis is a crucial field of research that involves detecting and understanding genetic variations occurring in cancer cells. Genomic sequencing is a technique used to determine the entire genome of a tissue or cell. In cancer research, this technology helps researchers identify which genes have mutated. Common genomic sequencing technologies include NGS (Next-Generation Sequencing) and single-cell sequencing, which are often used to analyze individual genes in cancer cells.
2.1.1 Next-Generation Sequencing (NGS) technology
Next-Generation Sequencing (NGS) is a high-throughput sequencing technology that has been widely applied in biological and medical research (Wu et al., 2019). The principle of NGS involves fragmenting DNA or RNA molecules into small segments, then sequencing these fragments in parallel, and finally assembling them into complete genomic or transcriptomic information (Figure 2).
|
Figure 2 principle of next generation sequencing (NGS) technology |
NGS technology boasts high throughput capabilities, enabling the simultaneous sequencing of a large number of DNA or RNA fragments. This allows it to generate vast amounts of data and quickly analyze entire genomes or transcriptomes. NGS technology employs parallel sequencing, which means sequencing multiple DNA or RNA fragments simultaneously. This increases sequencing speed, reduces costs, and makes sample processing more efficient. NGS can be used for various applications, including whole-genome sequencing, whole-exome sequencing, RNA sequencing, methylation sequencing, and ChIP sequencing. It is widely applied in genomics, transcriptomics, and epigenetics.
NGS technology plays a crucial role in cancer research, genetics research, pathogen identification, and drug development. It is used to detect gene mutations, discover new genes, analyze expression profiles, and uncover disease mechanisms. NGS technology holds great potential in personalized medicine and precision medicine. It can be used to diagnose diseases, guide treatment choices, monitor therapeutic efficacy, and predict disease risk. The large volume of data generated by NGS requires complex data analysis methods. The development of bioinformatics tools and algorithms is essential for extracting useful information from raw sequencing data.
2.1.2 Single-cell sequencing
Single-cell sequencing is a highly precise genomic technology used to analyze and sequence the genome, transcriptome, and epigenome of individual cells. Unlike traditional bulk sequencing methods, single-cell sequencing can reveal genetic and expression differences between individual cells and the cellular heterogeneity within a population.
Single-cell sequencing provides high-resolution information at the cellular level, uncovering genetic and expression differences between cells, which helps in understanding cellular heterogeneity. This technology can help researchers discover new cell types or subtypes that might be hidden in traditional sequencing. By tracking gene expression changes in individual cells, single-cell sequencing can aid in studying cellular development and differentiation processes. It can decompose a complex cell population into distinct subgroups, identifying key genes and biological characteristics within each subgroup.
Single-cell sequencing can be used to study the genetic and expression differences among different cell subpopulations within tumors, identify potential therapeutic targets, and understand tumor progression mechanisms. In immunology research, single-cell sequencing can reveal the diversity of immune cells, including T cells, B cells, macrophages, and their roles in infection and immune response. It can also be used in neuroscience to study different types of neurons in the brain and explore the mechanisms of neurological diseases. Single-cell sequencing helps understand embryonic development and organ formation processes, as well as the regulatory mechanisms of cell differentiation, playing a significant role in developmental biology. Additionally, it can analyze patient samples to understand individual disease differences and guide personalized treatment strategies.
2.2 Bioinformatics tools and databases
In cancer research and mutation analysis, bioinformatics tools and databases play a crucial role. These databases and tools provide important resources and support for mutation analysis, helping to uncover the biological and clinical significance of cancer mutations and guide the development of personalized treatment strategies.
2.2.1 Important mutation databases
The Cancer Genome Atlas (TCGA) is a database that includes multiple cancer types and provides extensive genomic data, including mutations, copy number variations, RNA expression, and clinical information. Researchers can find mutation information for various cancers here. The Catalogue of Somatic Mutations in Cancer (COSMIC) is a database containing mutation information from various cancers. It provides detailed mutation annotations and classifications, helping to study the function and impact of mutations.
ClinVar aggregates clinical information related to genetic variations, including the association of mutations with diseases. It is very useful for understanding the clinical significance of mutations. dbSNP is a database that contains information on various genetic variations, including single nucleotide polymorphisms (SNPs). Although it primarily focuses on genetic polymorphisms, it also includes mutation information. The Clinical Interpretations of Variants in Cancer (CIViC) is a database dedicated to organizing clinical interpretation information for cancer mutations, helping to understand the relationship between mutations and treatment responses.
2.2.2 Data analysis methods
To analyze whole genome or transcriptome data, researchers can use a series of bioinformatics tools and workflows, such as GATK (Genome Analysis Toolkit), Mutect, SAMtools, and others. Machine learning algorithms can be used to predict the functional impact, clinical relevance, and patient classification of mutations. Commonly used machine learning libraries include scikit-learn and XGBoost.
Mutation annotation tools, such as ANNOVAR, Variant Effect Predictor (VEP), and SnpEff, are used to annotate mutations and help determine their functional impacts (Dhanda et al., 2023). Variant frequency analysis is used to determine the frequency of mutations in populations, such as Exome Aggregation Consortium (ExAC) and Genome Aggregation Database (gnomAD). Mutation mapping tools, such as cBioPortal and Integrative Genomics Viewer (IGV), can visualize the distribution of mutations in the genome and the co-occurrence of mutations.
3 Clinical Applications of Cancer Mutation Analysis
3.1 Personalized treatment
In the field of cancer treatment, personalized treatment is a groundbreaking clinical strategy designed to tailor the most effective treatment plan based on each patient's unique genetic characteristics and tumor mutation status. Cancer mutation analysis plays a crucial role in personalized treatment, helping doctors select the most appropriate treatment methods, primarily including targeted therapy and immunotherapy.
3.1.1 Targeted therapy
Targeted therapy is a treatment approach based on specific molecular targets that intervenes in key signaling pathways essential for tumor cell growth, proliferation, or survival, achieving a more precise therapeutic effect. Cancer mutation analysis helps doctors identify specific target mutations present in the patient's tumor, enabling them to choose the most suitable targeted drugs. For example, in patients with EGFR mutations in non-small cell lung cancer, drugs such as Gefitinib and Erlotinib have been successfully used for targeted therapy, improving survival rates and quality of life. For melanoma patients with BRAF V600E mutations, BRAF inhibitors can be selected (Aleksakhina and Imyanitov, 2021). This personalized treatment strategy can enhance therapeutic efficacy, reduce unnecessary drug toxicity, and extend patient survival.
3.1.2 Immunotherapy
Immunotherapy is a treatment method that utilizes the patient's own immune system to combat cancer. Cancer mutation analysis can help determine whether the patient's tumor has a high tumor mutation burden (TMB), indicating a large number of mutations in the tumor. Tumors with high TMB are usually more easily recognized and attacked by the immune system. Therefore, for these patients, immune checkpoint inhibitors such as PD-1 and CTLA-4 inhibitors may be effective treatment options. Additionally, by analyzing the tumor's mutation burden and immune cell infiltration, it is possible to better predict the efficacy of immunotherapy, thereby achieving a more personalized treatment strategy.
3.2 Drug development
3.2.1 Drug screening and design
Drug screening and design are key applications of cancer mutation analysis in the field of drug development. In the initial stages of drug screening and design, the primary task is to identify potential therapeutic targets or molecules. Cancer mutation analysis plays a crucial role in this process by revealing key gene mutations associated with the occurrence and development of cancer. By analyzing the mutation profiles of different types of cancer patients, researchers can identify common pathogenic mutations or mutational pathways that can serve as potential therapeutic targets.
Once potential therapeutic targets are identified, researchers can conduct in vitro drug screening experiments. This typically involves applying known or potential anticancer drugs to tumor cell lines or animal models to evaluate their inhibitory effects on specific targets or mutations. These experiments can help determine which drugs have the best inhibitory effects on specific mutations or mutational pathways.
In addition to existing drug screening, drug development also involves the design and development of new drugs. Cancer mutation analysis provides valuable information for drug design. Researchers can design drug molecules with higher affinity and selectivity based on known mutation structures. These drugs may be targeted, capable of precisely intervening in specific mutations or mutational pathways, thereby reducing unnecessary side effects. After drug screening and design, candidate drugs need to undergo comprehensive evaluation, including preclinical and clinical trials. In these stages, the efficacy and safety of the drugs will be validated to ensure their feasibility in clinical treatment.
3.2.2 Drug resistance research
Drug resistance research is another important aspect of cancer mutation analysis in drug development and treatment. Drug resistance refers to the phenomenon where patients initially respond effectively to a therapeutic drug, but over time, cancer cells gradually lose sensitivity to the drug, leading to diminished or failed treatment efficacy.
The mechanisms of drug resistance are complex and may involve various factors. Cancer mutation analysis can help uncover the molecular and genetic changes leading to drug resistance (Gao et al., 2020). This includes the emergence of new mutations that alter drug targets or activate resistance-related pathways within the cells. Understanding these mechanisms is crucial for finding new strategies to combat resistance. Researchers aim to identify biomarkers that can be used for early detection of drug resistance. By analyzing cancer cells or patient samples, molecular features related to resistance, such as protein expression, gene expression, or mutation status, can be identified. These biomarkers can help predict whether a patient is likely to develop resistance, allowing for early adjustments to treatment plans.
Once resistance mechanisms are understood, researchers can develop corresponding treatment strategies. This may include the development of new drugs to overcome known resistance mechanisms or the adoption of combination therapy strategies to simultaneously target multiple resistance pathways. Additionally, optimizing drug dosages and treatment plans can help delay or mitigate the development of resistance. In clinical practice, monitoring patients for drug resistance is crucial. By regularly assessing patients' mutation status and treatment responses, doctors can detect signs of resistance early and adjust treatment plans as needed to prolong efficacy.
3.3 Early cancer diagnosis
Early cancer diagnosis is one of the key factors for successful cancer treatment, as treatment is usually easier and more successful when diagnosed early. The development of liquid biopsy and urine testing represents significant breakthroughs in the field of early cancer diagnosis, with the potential to improve detection rates while reducing patient discomfort and risk. However, these methods often require further research and validation to ensure their accuracy and reliability.
3.3.1 Liquid biopsy
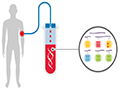
Liquid biopsy is a non-invasive diagnostic method that detects cancer by analyzing biomarkers in blood or other body fluids. These biomarkers can include circulating DNA, RNA, proteins, or cells. In cancer patients, cancer cells release these biomarkers into body fluids. By extracting a small amount of blood or other body fluid samples, laboratories can analyze the biomarkers to determine the presence of cancer. The advantage of liquid biopsy is that it is non-invasive, eliminating the need for tissue biopsies or imaging tests. This makes early diagnosis easier while reducing patient discomfort and risk. Liquid biopsy has been applied to various types of cancer, including lung cancer, breast cancer, and colorectal cancer (Figure 3).
|
Figure 3 Schematic diagram of liquid biopsy |
3.3.2 Urine testing
Similar to liquid biopsy, urine testing is a non-invasive method used for early cancer diagnosis. In this method, urine samples are used to detect specific biomarkers that may be associated with cancer. Urine testing is commonly used for screening cancers of the urinary system, such as bladder cancer. The advantage of urine testing is its convenience; patients can easily collect urine samples without needing special equipment or skills. This method can also be used for regular monitoring of patients and screening high-risk populations. However, the application of urine testing across different types of cancer is relatively limited, as different cancers may involve different biomarkers.
4 Success Stories
4.1 Development of alectinib
Alectinib is a targeted therapy drug for lung cancer patients, particularly those with ALK-positive non-small cell lung cancer. The development of Alectinib is one of the outstanding examples of cancer mutation analysis in drug development. In the past, treatment options for lung cancer patients were limited, but through mutation analysis, researchers discovered that certain lung cancer patients carry ALK gene mutations, providing opportunities for targeted therapy (Yin et al., 2022).
Alectinib is specifically designed to inhibit the abnormal ALK gene in patients. Through structural and functional analysis, researchers were able to precisely design a drug that interferes with the ALK signaling pathway. Alectinib showed high efficacy in clinical trials, significantly extending the survival of patients with ALK-positive non-small cell lung cancer. This provided strong support for the further development and approval of the drug. Alectinib eventually gained regulatory approval and was applied in clinical practice. This case emphasizes the critical role of mutation analysis in drug development and how these analyses can be translated into innovative cancer treatments.
4.2 Clinical application of osimertinib
Osimertinib is an effective drug for the personalized treatment of NSCLC patients, particularly those with the EGFR T790M mutation (Yin et al., 2022). The development of Osimertinib began with an in-depth study of the EGFR T790M mutation. This mutation is one of the main causes of resistance to EGFR inhibitors, but mutation analysis revealed that this mutation is a potent target for treatment.
By analyzing the structure and function of the mutation, scientists were able to design Osimertinib, a specific drug that precisely inhibits the EGFR T790M mutation and reduces drug resistance. Osimertinib showed high efficacy in clinical trials, especially for patients with the EGFR T790M mutation. This made Osimertinib a revolutionary lung cancer treatment, offering new hope to this subgroup of patients.
4.3 Breakthroughs in immunotherapy
Immunotherapy represents a significant breakthrough in cancer treatment, based on mobilizing the patient's immune system to fight cancer cells. Through mutation analysis, scientists can identify the mutation burden in a patient's tumor cells, i.e., the accumulation of mutations. There is a correlation between these mutation burdens and the patient's immune response. Mutation analysis revealed mutation antigens in patient tumors, providing crucial information for the development of targeted vaccines and immune checkpoint inhibitors. The clinical success of immunotherapy, such as the application of immune checkpoint inhibitors, has benefited an increasing number of patients. This groundbreaking treatment method has achieved significant success in various cancer types.
5 Summary and Outlook
This review delved into the key aspects of cancer mutation analysis, including types of mutations, molecular biology techniques, clinical applications, and success stories. Cancer mutations are diverse, including point mutations, deletions, insertions, and structural variations, providing multiple dimensions for cancer research. Genomic sequencing technologies, particularly next-generation sequencing (NGS), have become powerful tools for identifying and validating cancer mutations (Zhao et al., 2023). The development of data management and bioinformatics tools is crucial for cancer mutation analysis, helping researchers effectively store, analyze, and interpret vast amounts of biological information data. Personalized treatment and precision medicine are transforming the paradigm of cancer treatment, allowing doctors to provide the most suitable treatment plan for each patient.
Cancer mutation analysis holds broad potential and applications in drug development. By deeply studying the genetic variations in cancer, researchers can identify drug targets and develop more targeted anticancer drugs. Identifying mechanisms of drug resistance allows for early adjustment of treatment plans. Improving the efficiency of clinical trials ensures that drugs progress to market faster. Providing better treatment options for clinicians improves patient survival rates and quality of life. These applications will continue to highlight the importance of cancer mutation analysis in drug development, providing more opportunities for the discovery and development of new drugs.
Despite significant progress in cancer mutation analysis, challenges and issues remain to be addressed. With the continuous advancement of bioinformatics technologies, the volume of data generated is increasing rapidly, making data management and privacy issues particularly important. Future research needs to develop better data storage and sharing policies while ensuring patient privacy rights are protected. Cancer mutation analysis requires multidisciplinary collaboration, including experts from biology, medicine, bioinformatics, and pharmaceutical chemistry. Future research will emphasize interdisciplinary cooperation to more comprehensively understand the complexity of cancer mutations.
With continuous technological advancements, we can expect more innovations in cancer mutation analysis. Some emerging trends may include the widespread application of single-cell analysis to better understand mutations within heterogeneous tumors, more research on immunotherapy to provide additional treatment options for cancer patients, more complex data analysis methods and tools to handle large-scale biological information data. In conclusion, the future of cancer mutation analysis is full of promise and will continue to contribute to the personalization and precision of cancer treatment.
Conflict of Interest Disclosure
The author affirms that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Aleksakhina S.N., and Imyanitov E.N., 2021, Cancer therapy guided by mutation tests: current status and perspectives, International Journal of Molecular Sciences, 22(20): 10931.
https://doi.org/10.3390/ijms222010931
PMid:34681592 PMCid:PMC8536080
Dhanda S.K., Mahajan S., and Manoharan M., 2023, Neoepitopes prediction strategies: an integration of cancer genomics and immunoinformatics approaches, Briefings in Functional Genomics, 22(1): 1-8.
https://doi.org/10.1093/bfgp/elac041
PMid:36398967
Gao M., Yang T.T., Li G.L., Chen R., Liu H.C, Gao Q., Wan K.L., and Feng S.D.,2020, Analysis on drug resistance-associated mutations of multi-drug resistant Mycobacterium tuberculosis based on whole-genome sequencing in China, Zhonghua Liuxingbingxue Zazhi (Chinese Journal of Epidemiology), 41(5): 770-775.
Harrison P.T., Vyse S., and Huang P.H., 2020, Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer, In Seminars in Cancer Biology, 61: 167-179.
https://doi.org/10.1016/j.semcancer.2019.09.015
PMid:31562956 PMCid:PMC7083237
Hodge R.G., Schaefer A., Howard S.V., and Der C.J., 2020, RAS and RHO family GTPase mutations in cancer: twin sons of different mothers?, Critical Reviews in Biochemistry and Molecular Biology, 55(4): 386-407.
https://doi.org/10.1080/10409238.2020.1810622
PMid:32838579
Liu Z.C., Li Z.X., Zhang Y., Zhou T., Zhang J.Y., You W.C., Pan K.f., and Li W.Q., 2021, Journal of interpretation on the report of global cancer statistics 2020, Zhongliu Zonghe Zhiliao Dianzi Zazhi (Multidisciplinary Cancer Management (Electronic Version)), 7(2): 1-13.
Mattiuzzi C., and Lippi G., 2019, Current cancer epidemiology, Journal of Epidemiology and Global Health, 9(4): 217.
https://doi.org/10.2991/jegh.k.191008.001
PMid:31854162 PMCid:PMC7310786
Wu A.S., Liu X.N., Liu Y.H., Liu G., and Liu L., 2019, Application of second generation gene sequencing data management and big data platform in precision medicine, Zhongguo Shengwu Gongcheng Zazhi (China Biotechnology), 39(2): 101-111.
Yin Q., Guo T., Zhou Y., Sun L., Meng M., Ma L., and Wang X., 2022, Effectiveness of alectinib and osimertinib in a brain metastasized lung adenocarcinoma patient with concurrent EGFR mutations and DCTN1‐ALK fusion, Thoracic Cancer, 13(4): 637-642.
https://doi.org/10.1111/1759-7714.14291
PMid:34964276 PMCid:PMC8841708
Zhao X.M., Zhang Y.X., Ma T.H., and Hu Y.F., 2023, Advances in cancer diagnosis and treatment based on NGS technology, Yaoxue Jinzhan (Progress in Pharmaceutical Sciences), 47(6): 404-416.
(2).png)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Yong Cheng
Related articles
. Cancer
. Mutation analysis
. Drug development
. Personalized treatment
. Immunotherapy
Tools
. Post a comment